The Open Bioinformatics Foundation (OBF) Event Fellowship program aims to promote diverse participation at events promoting open-source bioinformatics software development and open science practices in the biological research community. Hetvi J, a PhD student at Imperial College London, was awarded an OBF Event Fellowship to attend BioHackathon Europe 2025.
Thanks to the Event Fellowship from the Open Bioinformatics Foundation (OBF), I had the opportunity to attend BioHackathon Europe 2025 located in the beautiful city of Bad Saarow, Germany. I’m currently a 3rd year PhD student in Biostatistics and my work focuses on human mitochondrial genetics. Specifically, I construct statistical models to test whether the presence or absence of somatic mitochondrial variants is associated with gene expression changes indicative of stress and aging. As part of my PhD, I work with single-cell -omics data, and run bioinformatics pipelines over high performance computing systems. My broader interests in the fields of open-source scientific computing and bioinformatics motivated me to participate in this hackathon.

Background on the Hackathon
This event was a 5 day Hackathon with 31 projects, attended by 170+ in person and 200+ online. The hackathon began with presentations of each project & we started working with our chosen teams on day 1. Every day, we had hacking sessions from 9am until 5:30pm & the evenings were followed by social events to facilitate networking between people.
During the hacking sessions, participants would work together with their project teams on assigned tasks & make progress by co-working with team members. On the last day, we had final presentations detailing work done and future plans from the diverse range of projects.

The projects are all in bioinformatics but spanning a variety of topics— everything from sustainable cluster computing to metadata standardization to building new pipelines. A full list of projects can be found here: https://biohackathon-europe.org/projects/
Brief information about my project
I was part of Project 4: Beyond Beacons- Establishing genomic background in European and international biobanks.
We had 5 in person members and 1 online member working collaboratively on this project. The broad goal of this project was to assess the effect of genomic context as defined by haploblocks on the penetrance of mendelian variants.
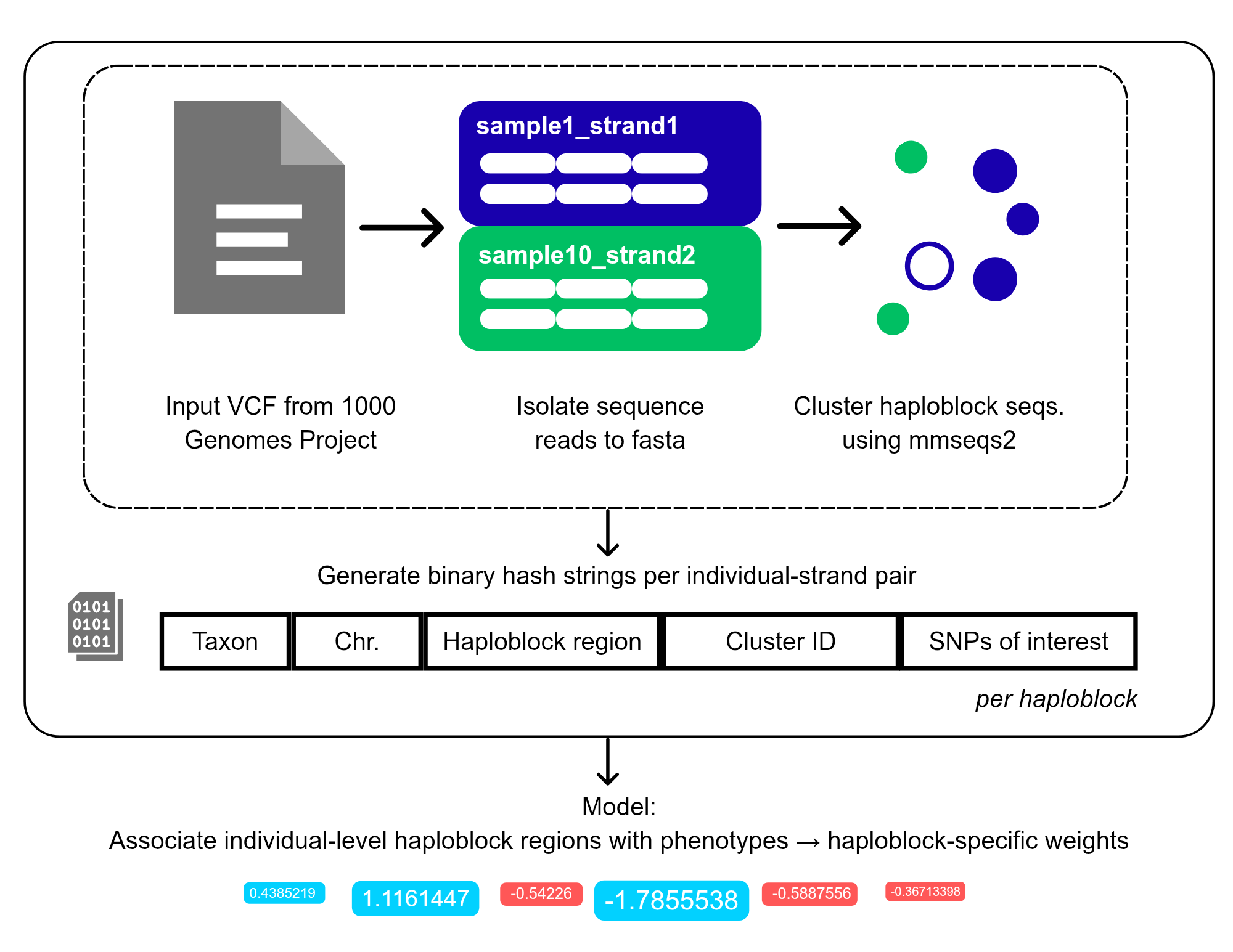
We segregated sequences from specific haploblocks that contain genes of interest and clustered them using mmseqs2 over 2600 individuals from the 1000 genomes project. Further, we developed a nomenclature system using binary strings to jointly-label the chromosomal, haploblock, cluster, and variant context per individual.
We also made progress on sketching statistical models that would help associate these per-individual hashes with phenotypes. I contributed to building the nomenclature rules, and to model development and the codebase. More information on the project can be found at our Github repository. We will be releasing a Docker pipeline and a pre-print based on our work soon!
Our Github: https://github.com/collaborativebioinformatics/Haploblock_Clusters_ElixirBH25

Social events
The hackathon had some amazing social events I took part in.

On Day 2, we had ‘Chain Reaction’ - an event where we made a creative obstacle course with all sorts of materials like wood or rubber ducks or even bubble machines to create a visual spectacle across 12 tables (the pictures will help!). A very unconventional but incredibly fun social activity- it was one of my favourite parts of the hackathon!

On Day 3, we had a mid-week progress update from each team through a Poster session. This was a great opportunity to learn about other projects in more detail and learn about their technical & organizational approaches to the hackathon.
Every day, we had walks at 6pm led by one of the hackathon organizers - which was a nice chance to see the lovely town of Baad Saarow with other participants.
Learning experiences
Even with my limited background in the topic of recombination & haploblocks— I was able to rapidly understand the project context & make meaningful contributions through collaborating with others & learning from them. It was also fun working collaboratively on a codebase.
Moreover, getting a chance to interact with people from the wider bioinformatics community across sub-disciplines was very valuable for me. It was fascinating to hear about the interesting and diverse backgrounds other participants came from— my learnings from these conversations will definitely guide me as I progress in my career.
Thanks again to OBF for enabling my participation in this project.
