The Open Bioinformatics Foundation (OBF) Event Fellowship program aims to promote diverse participation at events promoting open-source bioinformatics software development and open science practices in the biological research community. Fatemeh Mirzade, a PhD researcher at the University of Antwerp, was awarded an OBF Event Fellowship to attend the BioHackathon Europe 2025 (https://biohackathon-europe.org/2025).

My BioHackathon Experience: Contributing to Open Science in Proteomics
From the 3rd to 7th of November 2025, I had an incredible opportunity that shaped my early PhD journey: participating in BioHackathon Europe 2025 as part of Project #29 on proteomics quality control. Thanks to the Open Bioinformatics Foundation (OBF) Event Fellowship, I joined a team of bioinformaticians and proteomics researchers working to solve a critical challenge in our field.
🔗 Project #29 repository: https://github.com/MS-Quality-Hub/biohackathon2025 🔗 OBF on GitHub: https://github.com/OBF
Why I Joined
As someone just starting their PhD, I was both nervous and excited to dive into this collaborative coding event. Our project aimed to build automated quality control frameworks for public proteomics repositories like PRIDE, which contains over 31,000 datasets. The problem? Without standardized quality metrics, researchers can’t confidently reuse existing data, essentially flying blind when trying to assess if a dataset is suitable for their analysis.
My Contribution: Making Sense of Quality Metrics
I was assigned to create a formal taxonomy for QC metrics in mass spectrometry. When I first looked at the existing PSI-MS Controlled Vocabulary, I was overwhelmed. There were metrics scattered everywhere, categorized in ways that mixed fundamentally different information together.
Working with the team, I helped develop a seven-dimensional classification system that describes each metric along orthogonal axes:
- workflow stage
- analytical dimension
- information dependency
- measurement scope
- acquisition strategy
- quality interpretation
- metric value type
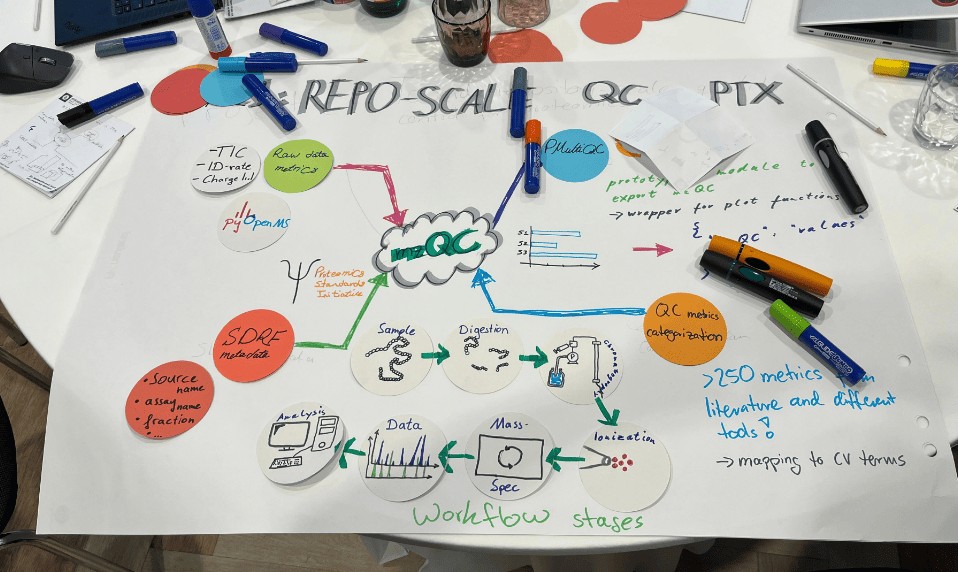
I compiled and classified 94 metrics from scientific literature, covering everything from chromatographic performance to identification confidence. For each one, I documented PSI-MS accession numbers, wrote descriptions, specified units, and noted applicability to different acquisition workflows. The work was tedious but deeply educational. By the end, I understood what “quality” actually means in proteomics.
What Open Source Contribution Feels Like
This was my first real experience contributing to open-source infrastructure that will be used globally. The spreadsheet I created is now feeding directly into updates to the PSI-MS Controlled Vocabulary. The team used my work to add new relationship types to the ontology, enabling tools to automatically understand what each metric means.
What struck me most was the collaborative nature of open science. I watched established researchers patiently explain concepts, saw consensus emerge through respectful debate, and learned that asking “stupid” questions often reveals important assumptions.
Meanwhile, other team members built incredible tools:
- an mzQC export function for pMultiQC
- an ID-free QC calculator using pyOpenMS
- a web-based validator for our classification system
These weren’t just internal prototypes but contributions to the broader proteomics ecosystem.
Beyond Technical Skills
Yes, I learned about mzQC formats, PSI-MS ontologies, and GitHub workflows. But the most valuable learning was about how open science actually functions. I experienced the generosity of researchers who take time to mentor newcomers, learned that technical decisions involve tradeoffs between elegance and practicality, and discovered that it’s okay to feel out of your depth because that’s where real learning happens.
Coming in as a fresh PhD student, I felt intimidated by fast-moving technical discussions and unfamiliar acronyms. But that initial disorientation became exactly what made the experience valuable. I’m leaving not just with new technical knowledge but with lasting connections to a community committed to collaborative infrastructure development.
Why This Matters
We’re building something that will help researchers worldwide make better use of public proteomics data. Imagine filtering datasets by quality before downloading terabytes, training machine learning models on curated high-quality data, or conducting meta-analyses that properly account for technical variation. That’s the future we’re working toward.
This hackathon showed me what open science looks like in practice: people from different backgrounds coming together to tackle shared challenges, contributing their expertise freely, and building infrastructure that lifts the entire field.
I’m grateful to the Open Bioinformatics Foundation for making my participation possible and excited to continue contributing throughout my PhD.
Want to Explore More?
❖ Full story on Medium: https://medium.com/@fatemehmirzadehsarcheshmeh/building-the-future-of-proteomics-quality-control-my-experience-at-biohackathon-europe-2025-8487191e170f
❖ Project repo: https://github.com/MS-Quality-Hub/biohackathon2025
❖ OBF GitHub: https://github.com/OBF